CaPS: Calcium Pattern Search Go to Bioinformatics-Hub
This program serarch for calcium binidng sites in proteins. Please input protein sequence here using FASTA format
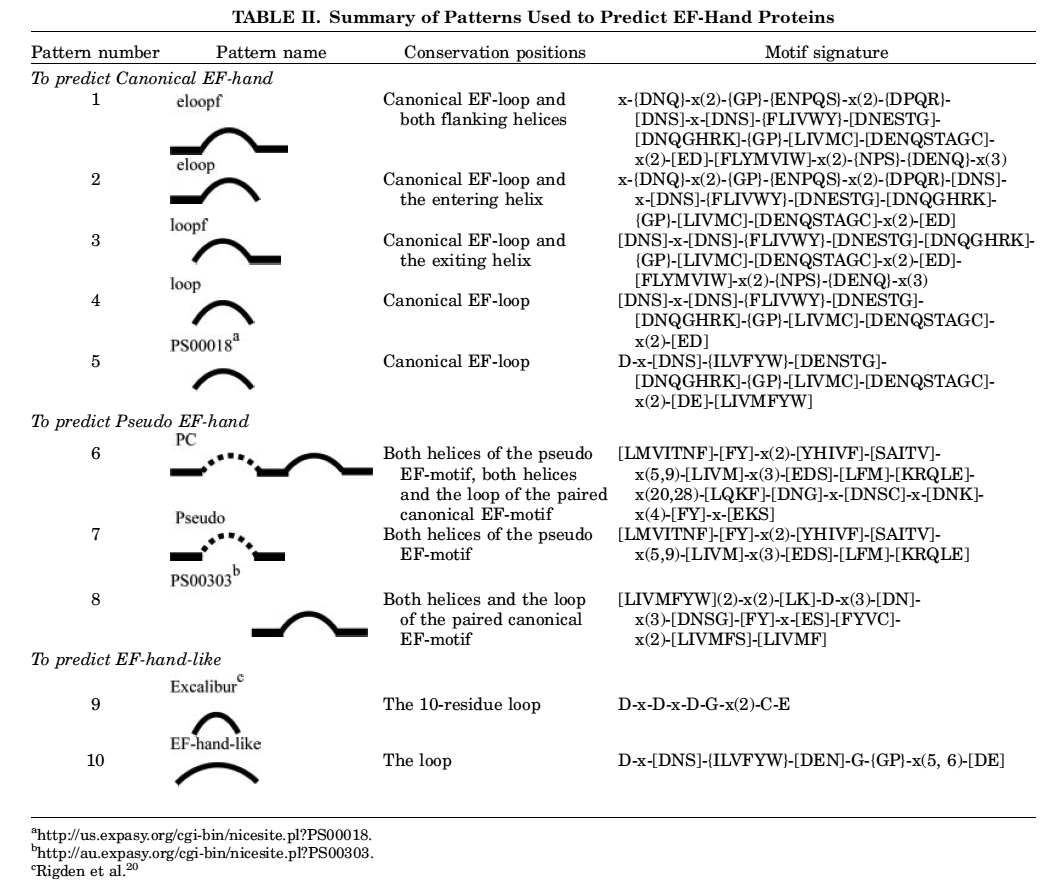
CaPS (Calcium Pattern Search) identifies Ca(II)-binding motifs in proteins based on unique patterns (or signatures) listed below. These patterns (or signatures) can be used to identify canonical EF-hand and pseudo EF-hand. We expect to significantly expand this list in the future to include non-contiguous (binding motif includes AA residues distant from each other in the primary structure) patterns. To check a sequence for the presence of a Ca(II)-binding signature, please past the protein sequence in the textarea and then click Search CAPS.
For more details, please read:
Zhou Y, Yang W, Kirberger M, Lee HW, Ayalasomayajula G, Yang JJ. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins. 2006 Nov 15;65(3):643-55.
Zhou Y, Xue S, Yang JJ. Calciomics: integrative studies of Ca2+-binding proteins and their interactomes in biological systems. Metallomics. 2013 Jan;5(1):29-42.
Zhou Y, Xue S, Chen Y, Yang JJ. Probing Ca2+-binding capability of viral proteins with the EF-hand motif by grafting approach. Methods Mol Biol. 2013;963:37-53
Y Chen, S Xue, Y Zhou, JJ Yang. Calciomics: prediction and analysis of EF-hand calcium binding proteins by protein engineering. Sci China Chem. 2010 Jan 1;53(1):52-60.